Bayesian modelling of summary statistics data
TL;DR. We develop Bayesian machine learning methods that excavate a latent structure or underlying joint models of summary statistics data.
What are summary statistics data?
There is no clear boundary to define that data are made available at the summary or observation level. Suppose the size of a data matrix/tensor $Z$ is smaller than the underlying sample size (number of observations). Each element in the data matrix $Z$ is generated as a result of some transformation of raw data. In that case, we often consider that the data $Z$ is observed at the summary level. Summary statistics data could be first and second-order moments (mean and variance), p-values, z-scores, effect sizes, and standard errors, etc. As the most well-known summary statistics data, we could think of the results of genome-wide association studies (GWAS).
Why do we bother summary statistics?
The foremost reason is that people feel pretty comfortable with sharing summary statistics. Unlike we have to deal with strict regulations to access individual-level data, a user (researcher) may find easily-downloadable GWAS summary statistics vectors from a public database. For the other party, those who provide data, there is almost no need to anonymize it since each data point does not correspond to an individual. For most clinical and public health studies, research groups share their final results as a giant summary statistics table. Other researchers gain essential knowledge by using them in subsequent research.
Great, but it sounds like everything is done and remains nothing to be done. I just said that summary statistics data are often final to some studies. Yes, to some people, but not to those who care about a generative model involving lots of parameters. Most summary data only investigate relationships between one variable with the other variable, but not many to many. Only a set of marginal probabilities, but not the joint probability of all. Can we recover a joint, multivariate model from a set of univariate models? Taking it further, can we estimate a joint probability of multiple studies conducted independently by different groups by combining summary data collected from each of them?
Summary-based regression modelling
Zhu and Stephens (2017) PMID: 29399241 proposed an interesting idea, termed regression with summary statistics (RSS) model. RSS recovers the high-dimensional parameters of a multivariate (linear) regression model, highlighting a correspondence between the multivariate and univariate association statistics.
We also found the RSS type of modelling can be effective enough to capture causal SNPs in high-dimensional variable selection problems. Here, we show simulation results on the reference panel genotype matrix ( the 1000 genomes project; on chromosome 1, European samples, n=503). We implemented a suite of related summary-based methods zQTL.
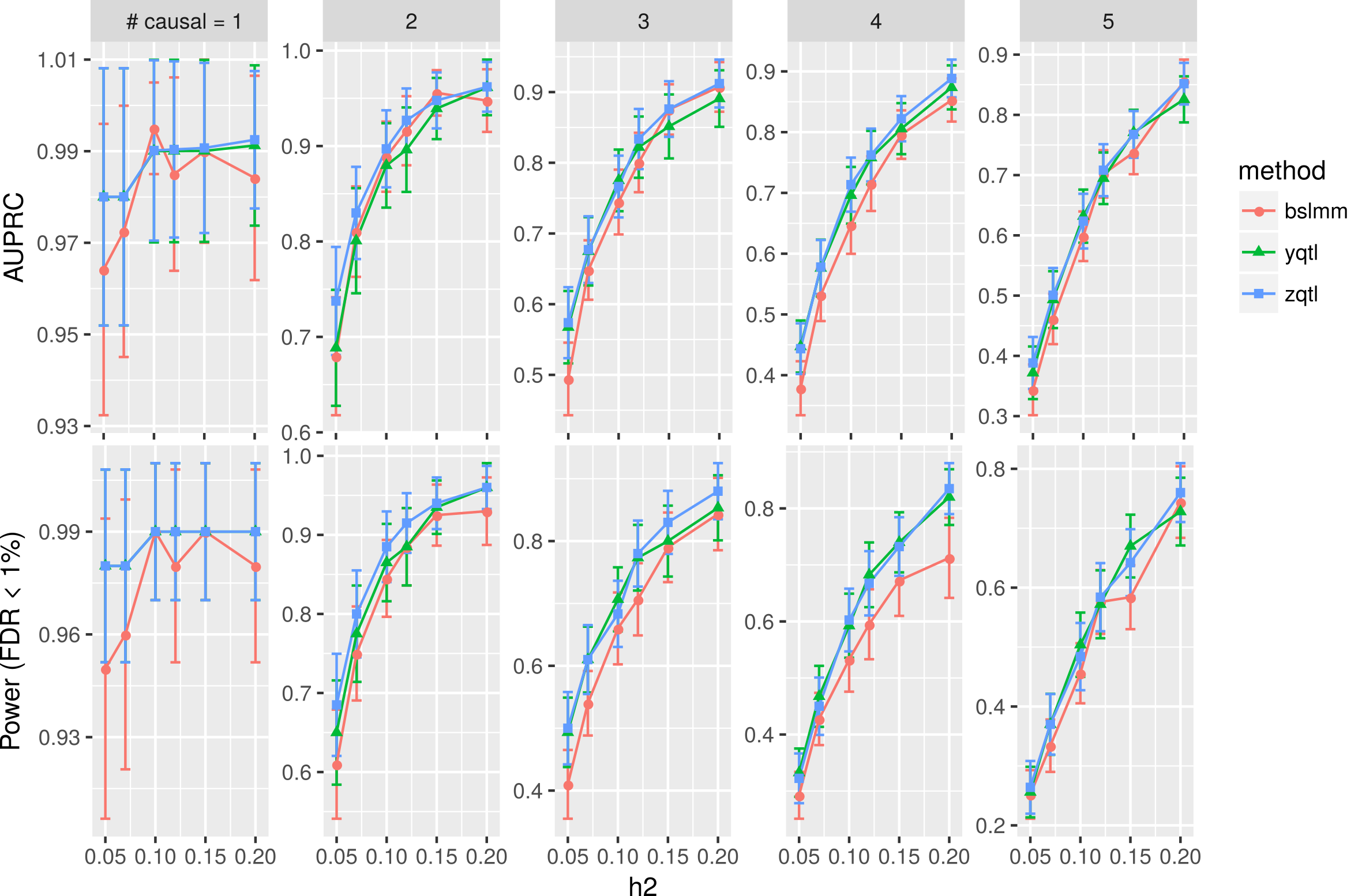
AUPRC: Area Under Precision Recall Curve. Column panels: simulations varying the number of causal SNPs. h2: the proportion of phenotypic variance explained by causal SNPs. BSLMM: Bayesian sparse linear mixed effect model; yqtl: a sparse regression model with spike-slab prior, trained by stochastic variational inference; zqtl: a summary-based multivariate regression modelling.
Summary statistics-based machine learning
Our next challenge is to widen the scope of computational biology, genomics applications that can be benefited from summary-based learning methods. About a decade ago, the ML community also realized the value of exploiting the spectral structure of the summary data matrix/tensor. Hsu et al. (2012) Hsu+Kakade+Zhang beautifully demonstrates an optimal state of a hidden Markov model can be easily recovered by the spectral decomposition of marginal probability matrices.