Variational inference of zQTL regression
fit.zqtl.RdVariational inference of zQTL regression
fit.zqtl(effect, effect.se, X)Arguments
- effect
Marginal effect size matrix (SNP x trait)
- effect.se
Marginal effect size standard error matrix (SNP x trait)
- X
Design matrix (reference Ind x SNP)
- n
sample size of actual data (will ignore if n = 0)
- A
Annotation matrix (SNP x annotations; default: NULL)
- multi.C
multivariate SNP confounding factors (SNP x confounder; default: NULL)
- univar.C
univariate SNP confounding factors (SNP x confounder; default: NULL)
- factored
Fit factored QTL model (default: FALSE)
- options
A list of inference/optimization options.
- do.hyper
Hyper parameter tuning (default: FALSE)
- do.rescale
Rescale z-scores by standard deviation (default: FALSE)
- tau
Fixed value of tau
- pi
Fixed value of pi
- tau.lb
Lower-bound of tau (default: -10)
- tau.ub
Upper-bound of tau (default: -4)
- pi.lb
Lower-bound of pi (default: -4)
- pi.ub
Upper-bound of pi (default: -1)
- tol
Convergence criterion (default: 1e-4)
- gammax
Maximum precision (default: 1000)
- rate
Update rate (default: 1e-2)
- decay
Update rate decay (default: 0)
- jitter
SD of random jitter for mediation & factorization (default: 0.1)
- nsample
Number of stochastic samples (default: 10)
- vbiter
Number of variational Bayes iterations (default: 2000)
- verbose
Verbosity (default: TRUE)
- k
Rank of the factored model (default: 1)
- svd.init
initialize by SVD (default: TRUE)
- right.nn
non-negativity in factored effect (default: FALSE)
- mu.min
mininum non-negativity weight (default: 0.01)
- print.interv
Printing interval (default: 10)
- nthread
Number of threads during calculation (default: 1)
- eigen.tol
Error tolerance in Eigen decomposition (default: 0.01)
- eigen.reg
Regularization of Eigen decomposition (default: 0.0)
- do.stdize
Standardize (default: TRUE)
- out.residual
estimate residual z-scores (default: FALSE)
- do.var.calc
variance calculation (default: FALSE)
- nboot
Number of bootstraps (default: 0)
- nboot.var
Number of bootstraps for variance estimation (default: 100)
- scale.var
Scaled variance calculation (default: FALSE)
- min.se
Minimum level of SE (default: 1e-4)
- rseed
Random seed
Value
a list of variational inference results.
param: sparse genetic effect size (theta, theta.var, lodds)
param.left: the left factor for the factored effect
param.right: the left factor for the factored effect
conf.multi: association with multivariate confounding variables
conf.uni: association with univariate confounding variables
resid: residuals
gwas.clean: cleansed version of univariate GWAS effects
var: variance decomposition results
llik: log-likelihood trace over the optimization
Details
Estimate true effect matrix from marginal effect sizes and standard errors (Hormozdiari et al., 2015; Zhu and Stephens, 2016): $$\mathbf{Z}_{t} \sim \mathcal{N}\!\left(R E^{-1} \boldsymbol{\theta}_{t}, R\right)$$ where R is \(p \times p\) LD / covariance matrix; E is expected squared effect size matrix (\(\textsf{se}[\boldsymbol{\theta}_{t}^{\textsf{marg}}] + n^{-1} \langle \boldsymbol{\theta}_{t}^{\textsf{marg}} \rangle^{2}\) matrix, diagonal); \(\mathbf{z}_{t}\) is \(p \times 1\) z-score vector of trait \(t\), or \(\mathbf{z}_{t} = \boldsymbol{\theta}_{t}^{\textsf{marg}}/ \textsf{se}[\boldsymbol{\theta}_{t}^{\textsf{marg}}]\).
In basic zQTL model, spasrse parameter matrix, \(\theta\) was modeled with spike-slab prior. We carry out posterior inference by variational inference with surrogate distribution first introduced in Carbonetto and Stephens (2012):
$$q(\theta|\alpha,\beta,\gamma) = \alpha \mathcal{N}\!\left(\beta,\gamma^{-1}\right)$$
We reparameterized \(\alpha = \boldsymbol{\sigma}\!\left(\pi + \delta\right)\), and \(\gamma = \gamma_{\textsf{max}}\boldsymbol{\sigma}\!\left(- \tau + \lambda \right)\) for numerical stability.
In factored zQTL model, we decompose sparse effect: $$\boldsymbol{\theta}_{t} = \boldsymbol{\theta}^{\textsf{left}} \boldsymbol{\theta}_{t}^{\textsf{right}}$$
Examples
###########################
## A simple zQTL example ##
###########################
n = 500
p = 2000
m = 1
set.seed(1)
.rnorm <- function(a, b) matrix(rnorm(a * b), a, b)
X = .rnorm(n, p)
Y = matrix(0, n, m)
h2 = 0.25
c.snps = sample(p, 3)
theta = .rnorm(3, m) * sqrt(h2 / 3)
Y = X[, c.snps, drop=FALSE] %*% theta + .rnorm(n, m) * sqrt(1 - h2)
library(dplyr)
library(tidyr)
qtl.tab = calc.qtl.stat(X, Y)
xy.beta = qtl.tab %>%
dplyr::select(x.col, y.col, beta) %>%
tidyr::spread(key = y.col, value = beta) %>%
(function(x) matrix(x[1:p, seq(2, m+1)], ncol = 1))
xy.se = qtl.tab %>%
dplyr::select(x.col, y.col, se) %>%
tidyr::spread(key = y.col, value = se) %>%
(function(x) matrix(x[1:p, seq(2, m + 1)], ncol = 1))
out = fit.zqtl(xy.beta, xy.se, X,
vbiter = 3000,
gammax = 1e3,
pi = 0,
eigen.tol = 1e-2,
do.var.calc = TRUE,
scale.var = TRUE)
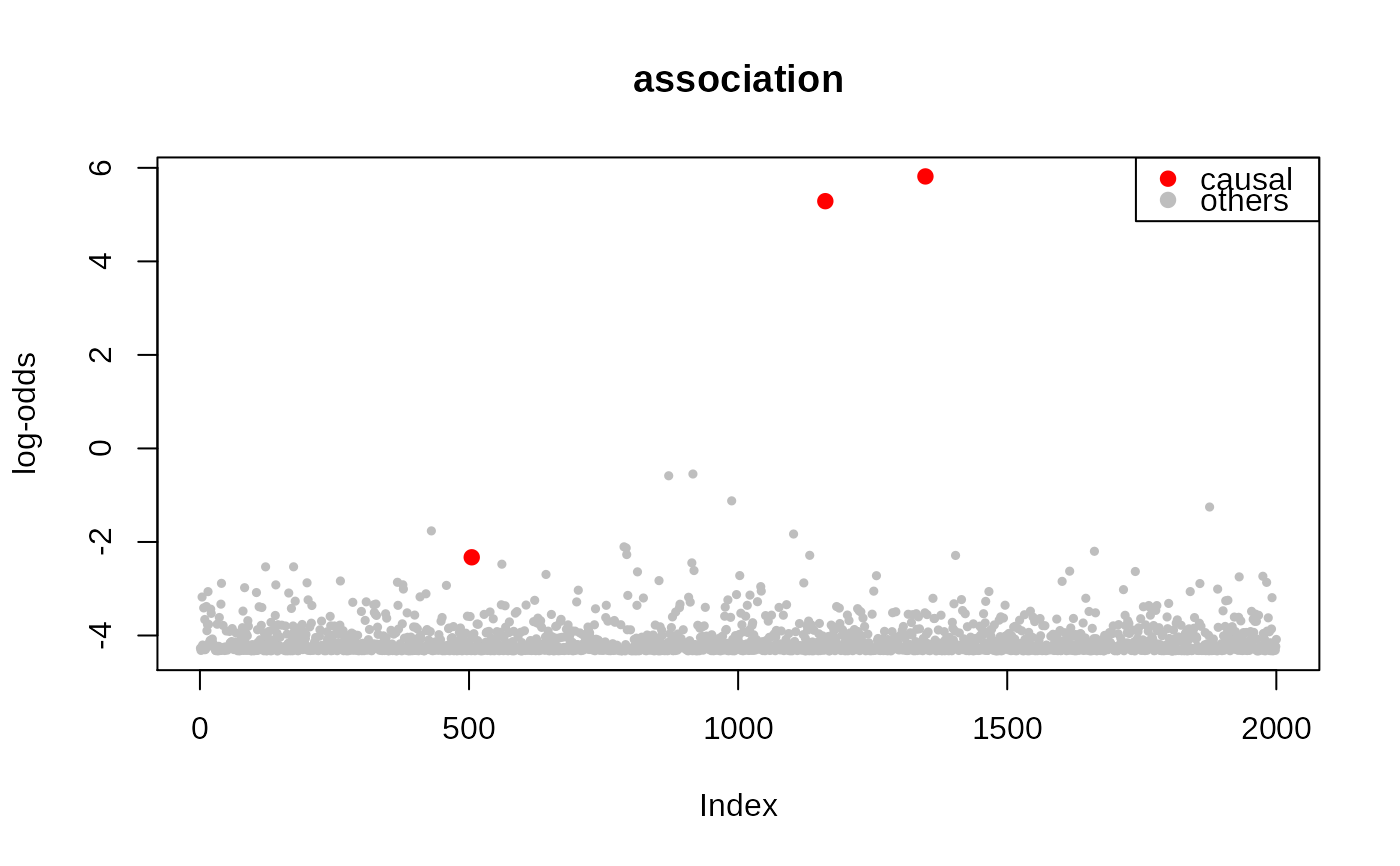
plot(out$param$lodds, main = 'association', pch = 19, cex = .5, col = 'gray', ylab = 'log-odds')
points(c.snps, out$param$lodds[c.snps], col = 'red', pch = 19)
legend('topright', legend = c('causal', 'others'), pch = 19, col = c('red','gray'))
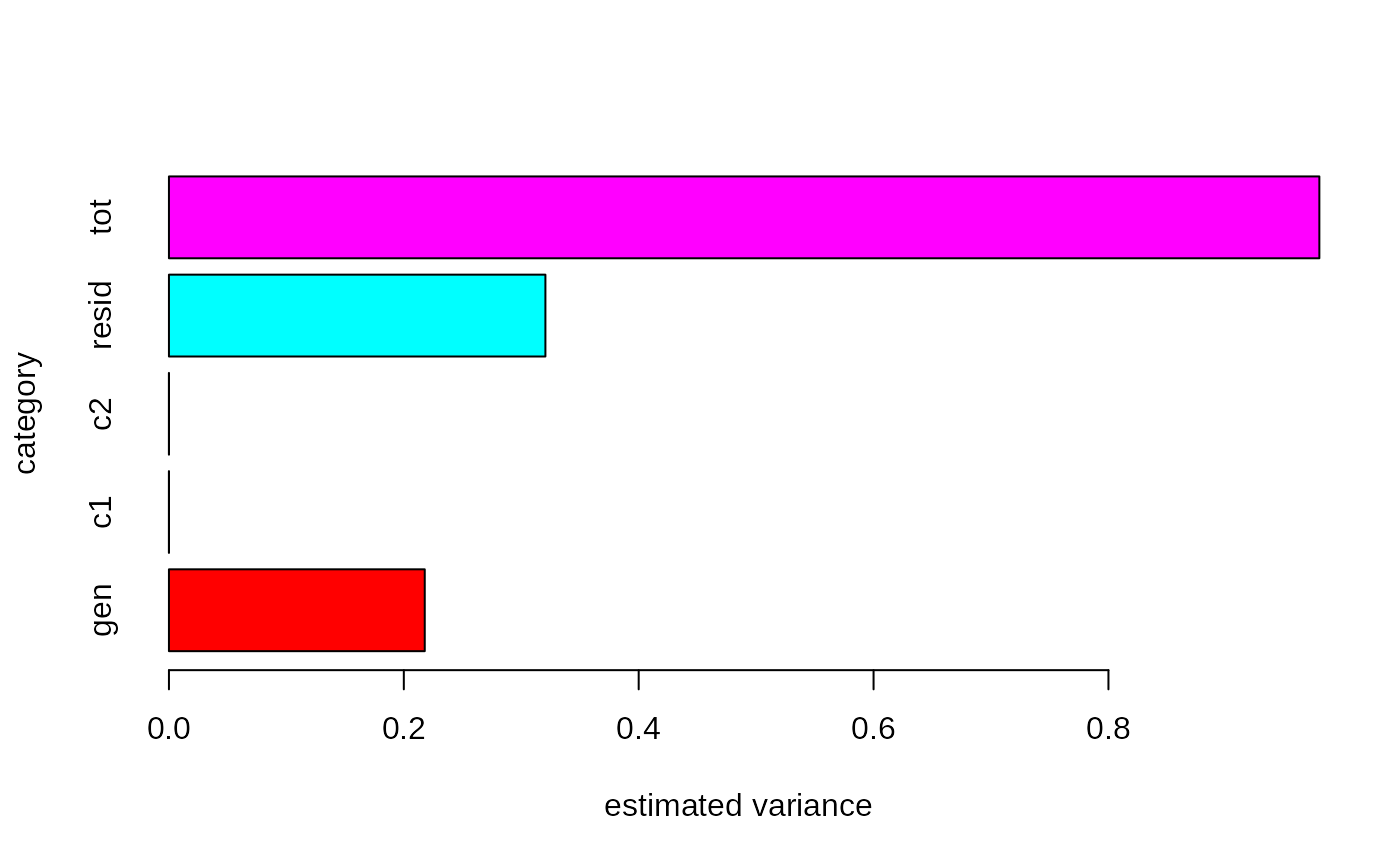
 .var = c(out$var$param$mean,
out$var$conf.mult$mean,
out$var$conf.uni$mean,
out$var$resid$mean,
out$var$tot)
barplot(height = 10^(.var),
col = 2:6,
names.arg = c('gen', 'c1', 'c2', 'resid', 'tot'),
horiz = TRUE,
ylab = 'category',
xlab = 'estimated variance')
.var = c(out$var$param$mean,
out$var$conf.mult$mean,
out$var$conf.uni$mean,
out$var$resid$mean,
out$var$tot)
barplot(height = 10^(.var),
col = 2:6,
names.arg = c('gen', 'c1', 'c2', 'resid', 'tot'),
horiz = TRUE,
ylab = 'category',
xlab = 'estimated variance')