Variational inference of zQTL mediation
fit.med.zqtl.RdVariational inference of zQTL mediation
fit.med.zqtl(effect, effect.se, effect.m, effect.m.se, X.gwas)Arguments
- effect
Marginal effect size matrix (SNP x trait)
- effect.se
Marginal effect size standard error matrix (SNP x trait)
- effect.m
Marignal genetic effects of mediators (SNP x mediator)
- effect.m.se
SE of the marginal genetic effects (SNP x mediator)
- X.gwas
Design matrix (reference Ind.GWAS x SNP)
- X.med
Design matrix (reference Ind.MED x SNP)
- n
sample size of GWAS data (will ignore if n = 0)
- n.med
sample size of mediation data (will ignore if n.med = 0)
- multi.C
SNP confounding factors (SNP x confounder; default: NULL)#'
- factored
Fit factored model (default: FALSE)
- options
A list of inference/optimization options.
- multivar.mediator
Multivariate mediator QTL effect (default: FALSE)
- de.factorization
Estimate direct effect by joint factorization (default: TRUE)
- factorization.model
Factorization model; 0 = ind x factor, 1 = eigen x factor (default: 0)
- num.strat.size
Size of stratified sampling (default: 2)
- do.direct.estimation
Estimate direct effect (default: TRUE)
- do.finemap.direct
Fine-map direct effect SNPs (default: FALSE)
- nboot
Number of bootstraps followed by finemapping (default: 0)
- nboot.var
Number of bootstraps for variance estimation (default: 100)
- scale.var
Scaled variance calculation (default: FALSE)
- do.var.calc
variance calculation (default: FALSE)
- num.duplicate.sample
Duplicate number of independent components (default: 1)
- num.conditional
number of conditional models
- submodel.size
size of each conditional model
- do.hyper
Hyper parameter tuning (default: FALSE)
- do.rescale
Rescale z-scores by standard deviation (default: FALSE)
- tau
Fixed value of tau
- pi
Fixed value of pi
- tau.lb
Lower-bound of tau (default: -10)
- tau.ub
Upper-bound of tau (default: -4)
- pi.lb
Lower-bound of pi (default: -4)
- pi.ub
Upper-bound of pi (default: -1)
- tol
Convergence criterion (default: 1e-4)
- gammax
Maximum precision (default: 1000)
- rate
Update rate (default: 1e-2)
- decay
Update rate decay (default: 0)
- jitter
SD of random jitter for mediation & factorization (default: 0.01)
- nsample
Number of stochastic samples (default: 10)
- vbiter
Number of variational Bayes iterations (default: 2000)
- verbose
Verbosity (default: TRUE)
- k
Rank of the factored model (default: 1)
- svd.init
initialize by SVD (default: TRUE)
- print.interv
Printing interval (default: 10)
- nthread
Number of threads during calculation (default: 1)
- eigen.tol
Error tolerance in Eigen decomposition (default: 0.01)
- eigen.reg
Regularization of Eigen decomposition (default: 0.0)
- do.stdize
Standardize (default: TRUE)
- out.residual
estimate residual z-scores (default: FALSE)
- min.se
Minimum level of SE (default: 1e-4)
- rseed
Random seed
Value
a list of variational inference results.
param.mediated: parameters for the mediated effects
param.unmediated: parameters for the unmediated effects
param.intercept: parameters for the intercept effect
param.covariate: parameters for the multivariate covariate
param.covariate.uni: parameters for the univariate covariate
bootstrap: bootstrapped mediated parameters (if nboot > 0)
llik: log-likelihood trace over the optimization
Details
Mediation analysis
Examples
library(dplyr)
#>
#> Attaching package: ‘dplyr’
#> The following objects are masked from ‘package:stats’:
#>
#> filter, lag
#> The following objects are masked from ‘package:base’:
#>
#> intersect, setdiff, setequal, union
n = 500
p = 1000
n.genes = 10
h2 = 0.8
n.causal.direct = 1
n.causal.qtl = 3
n.causal.gene = 2
set.seed(13)
.rnorm <- function(a, b) matrix(rnorm(a * b), a, b)
X.raw = sapply(1:p, function(j) {
f = runif(1, min = 0.1, max = 0.9)
rbinom(n, 2, f)
})
X = scale(X.raw)
c.qtls = matrix(sapply(1:n.genes, function(j) matrix(sample(p, n.causal.qtl))), nrow = n.causal.qtl)
theta.snp.gene = .rnorm(n.causal.qtl, n.genes) / n.causal.qtl
gene.expr = lapply(1:dim(c.qtls)[2], function(j) X[, c.qtls[,j], drop = FALSE] %*% theta.snp.gene[, j, drop = FALSE] + 0.5 * rnorm(n) )
gene.expr = do.call(cbind, gene.expr)
theta.med = sample(c(-1,1), n.causal.gene, TRUE)
causal.direct = sample(p, n.causal.direct)
theta.dir = matrix(rnorm(n.causal.direct), n.causal.direct, 1) / n.causal.direct
y = gene.expr[,1:n.causal.gene,drop = FALSE] %*% theta.med # mediation
y = y + X[, causal.direct, drop = FALSE] %*% theta.dir # direct
y = y + rnorm(n) * c(sqrt(var(y) * (1/h2 - 1)))
eqtl.tab = calc.qtl.stat(X, gene.expr)
gwas.tab = calc.qtl.stat(X, y)
xg.beta = eqtl.tab %>%
dplyr::select(x.col, y.col, beta) %>%
tidyr::spread(key = y.col, value = beta) %>%
(function(x) as.matrix(x[1:p, -1]))
xg.se = eqtl.tab %>%
dplyr::select(x.col, y.col, se) %>%
tidyr::spread(key = y.col, value = se) %>%
(function(x) as.matrix(x[1:p, -1]))
xy.beta = gwas.tab %>%
dplyr::select(x.col, y.col, beta) %>%
tidyr::spread(key = y.col, value = beta) %>%
(function(x) as.matrix(x[1:p, -1]))
xy.se = gwas.tab %>%
dplyr::select(x.col, y.col, se) %>%
tidyr::spread(key = y.col, value = se) %>%
(function(x) as.matrix(x[1:p, -1]))
vb.opt = list(nsample = 10,
vbiter = 3000,
rate = 1e-2,
gammax = 1e3,
do.stdize = TRUE,
pi = -0, tau = -4,
do.hyper = FALSE,
eigen.tol = 1e-2,
tol = 1e-8,
verbose = TRUE,
print.interv = 200,
multivar.mediator = FALSE,
nthread = 8)
med.out = fit.med.zqtl(effect = xy.beta,
effect.se = xy.se,
effect.m = xg.beta,
effect.m.se = xg.se,
X.gwas = X,
options = vb.opt)
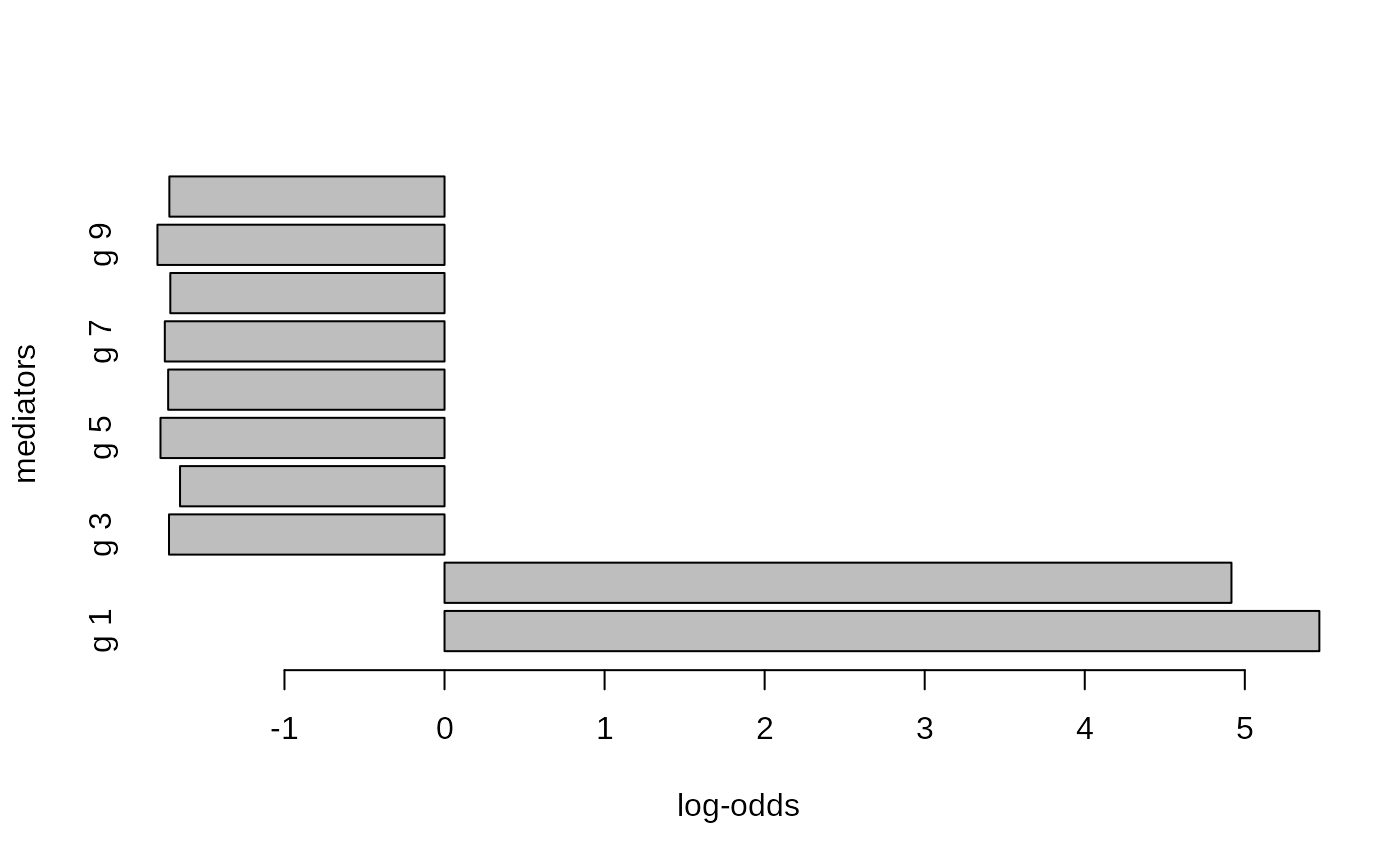
barplot(height = med.out$param.mediated$lodds[, 1],
names.arg = paste('g',1:n.genes),
horiz = TRUE, xlab = 'log-odds', ylab = 'mediators')